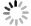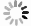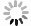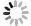Down-regulating XBP1s alleviates hypoxia/reoxygenation injury of renal tubular epithelial cells by inhibiting ITPR-mediated mitochondrial dysfunction
-
摘要:
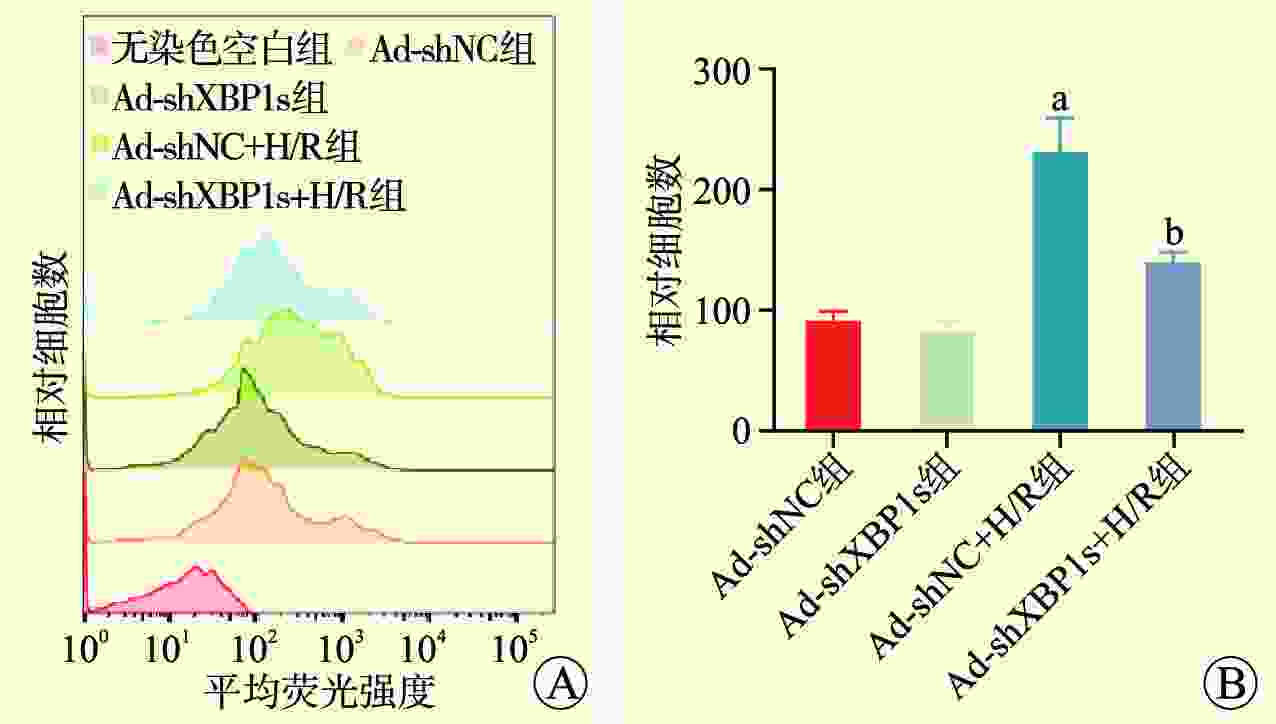
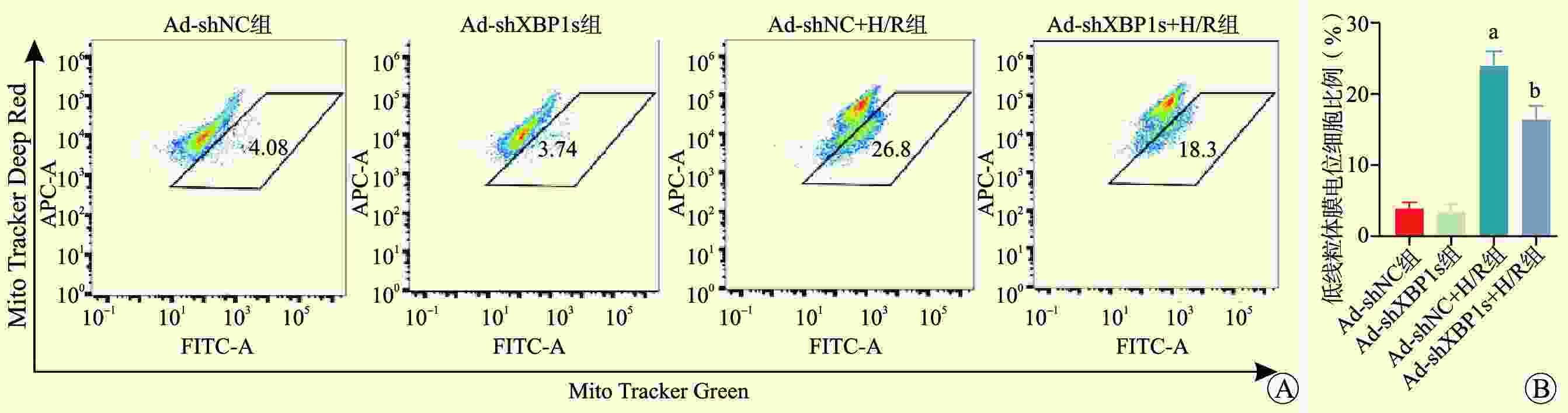
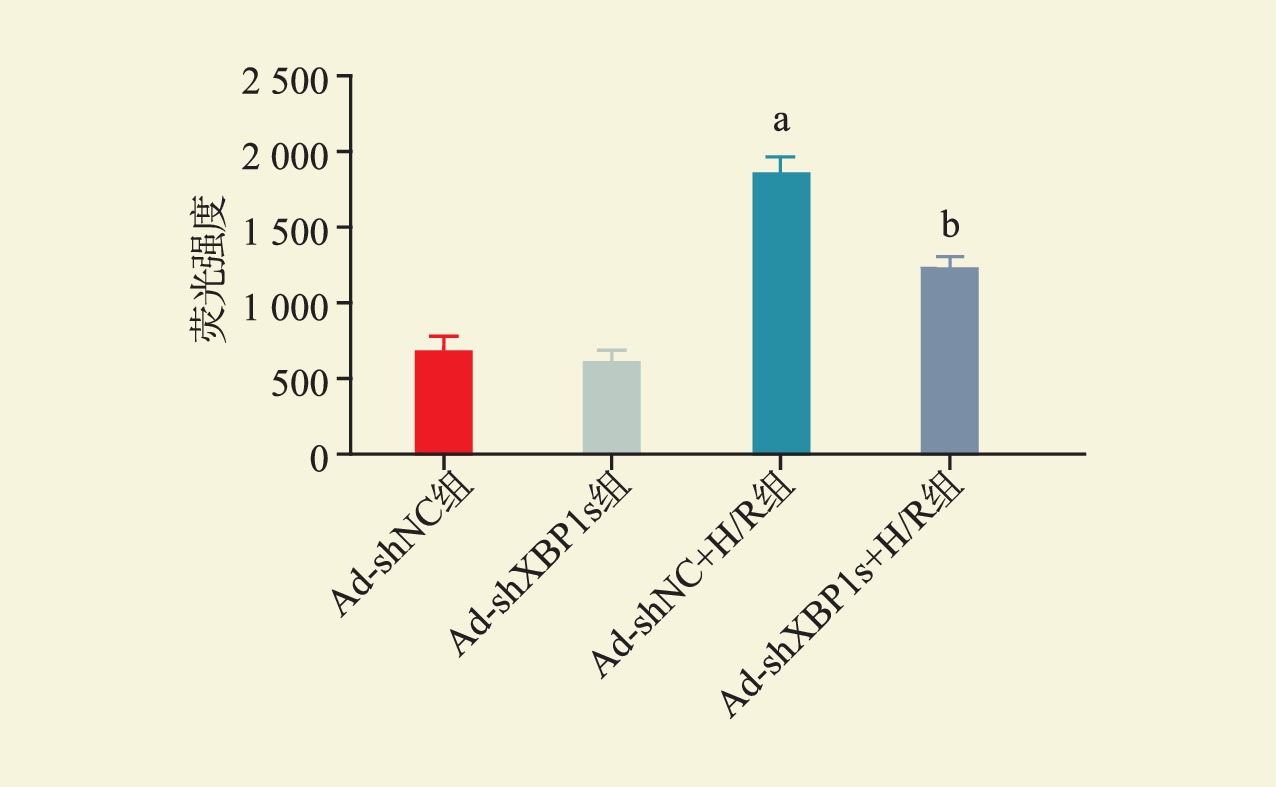
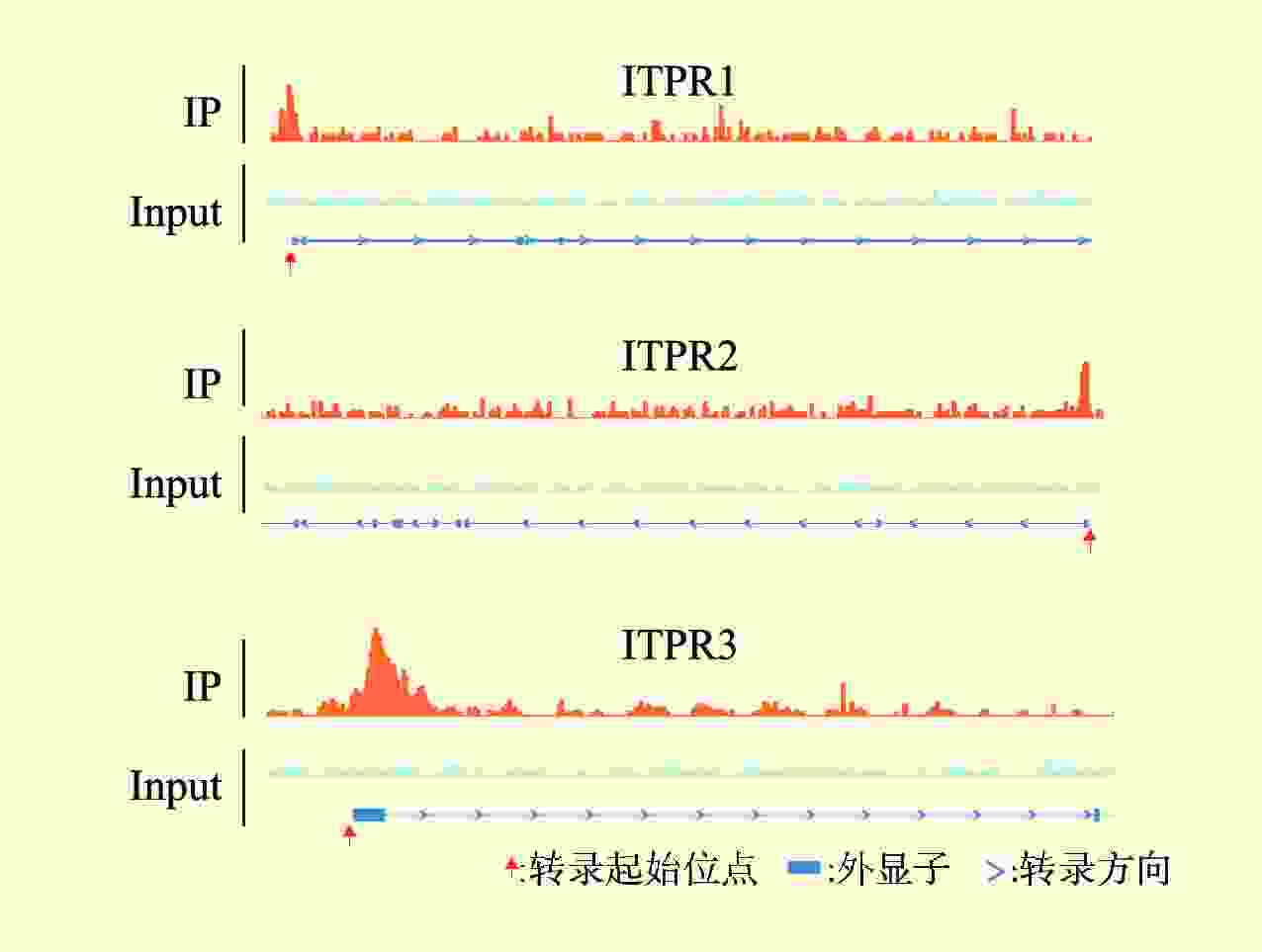
目的 探讨剪接型X盒结合蛋白1(XBP1s)对小鼠肾小管上皮细胞缺氧/复氧(H/R)损伤的影响及其作用机制。 方法 将小鼠肾小管上皮细胞分为腺病毒阴性对照组(Ad-shNC组)、靶向沉默XBP1s腺病毒组(Ad-shXBP1s组)、Ad-shNC+H/R组、Ad-shXBP1s+H/R组。检测各组细胞凋亡水平、线粒体活性氧活性、线粒体膜电位及线粒体钙离子水平。使用染色质免疫共沉淀测序(ChIP-seq)分析XBP1s调控肌醇1,4,5-三磷酸受体(ITPR)家族的结合位点。检测各组XBP1s和ITPR家族信使RNA(mRNA)和蛋白表达水平。 结果 与Ad-shNC组比较,Ad-shNC+H/R组细胞凋亡水平更高,线粒体活性氧水平升高,线粒体膜电位降低,线粒体钙离子水平升高;与Ad-shNC+H/R组比较,Ad-shXBP1s+H/R组细胞凋亡水平较低,线粒体活性氧水平下降,线粒体膜电位升高,线粒体钙离子水平降低(均为P<0.05)。与Ad-shNC组比较,Ad-shXBP1s组XBP1s、ITPR1、ITPR2和ITPR3 mRNA和蛋白相对表达量降低(均为P<0.05)。与Ad-shNC组相比,Ad-shNC+H/R组XBP1s、ITPR1、ITPR2和ITPR3蛋白相对表达量升高;与Ad-shNC+H/R组相比,Ad-shXBP1s+H/R组XBP1s、ITPR1、ITPR2和ITPR3蛋白相对表达量下降(均为P<0.05)。ChIP-seq结果显示,XBP1s能够结合ITPR1的启动子和外显子、ITPR2外显子和ITPR3外显子。 结论 XBP1s可能通过直接调控ITPR转录和翻译而影响线粒体相关的内质网膜结构功能,下调XBP1s能够抑制ITPR表达,改善线粒体损伤。 -
关键词:
- 器官移植 /
- 缺血-再灌注损伤 /
- 剪接型X盒结合蛋白1 /
- 肌醇1,4,5-三磷酸受体 /
- 线粒体损伤 /
- 内质网应激 /
- 线粒体相关的内质网膜 /
- 钙超载
Abstract:Objective To evaluate the effect of spliced X-box binding protein 1 (XBP1s) on hypoxia/reoxygenation (H/R) injury of mouse renal tubular epithelial cells and unravel underlying mechanism. Methods Mouse renal tubular epithelial cells were divided into adenovirus negative control group (Ad-shNC group), targeted silencing XBP1s adenovirus group (Ad-shXBP1s group), Ad-shNC+H/R group and Ad-shXBP1s+H/R group. The apoptosis level, mitochondrial reactive oxygen activity, mitochondrial membrane potential and mitochondrial calcium ion level were detected in each group. Chromatin immunocoprecipitation followed by sequencing (ChIP-seq) was employed to analyze the binding sites of XBP1s in regulating the inositol 1,4,5-trisphosphate receptor (ITPR) family. The expression levels of XBP1s and ITPR family messenger RNA (mRNA) and protein were determined in each group. Results Compared with the Ad-shNC group, the apoptosis level was higher, mitochondrial reactive oxygen species level was increased, mitochondrial membrane potential was decreased and mitochondrial calcium ion level was elevated in the Ad-shNC+H/R group. Compared with the Ad-shNC+H/R group, the apoptosis level was lower, mitochondrial reactive oxygen species level was decreased, mitochondrial membrane potential was elevated, and mitochondrial calcium ion level was decreased in the Ad-shXBP1s+H/R group (all P<0.05). Compared with the Ad-shNC group, relative expression levels of XBP1s, ITPR1, ITPR2 and ITPR3 mRNAs and proteins were down-regulated in the Ad-shXBP1s group (all P<0.05). Compared with the Ad-shNC group, relative expression levels of XBP1s, ITPR1, ITPR2 and ITPR3 proteins were up-regulated in the Ad-shNC+H/R group. Compared with the Ad-shNC+H/R group, relative expression levels of XBP1s, ITPR1, ITPR2 and ITPR3 were down-regulated in the Ad-shXBP1s+H/R group (all P<0.05). ChIP-seq results showed that XBP1s could bind to the promoter and exon of ITPR1, the exon of ITPR2, and the exon of ITPR3. Conclusions XBP1s may affect mitochondria-associated endoplasmic reticulum membrane structure and function by directly regulating ITPR transcription and translation. Down-regulating XBP1s may inhibit ITPR expression and mitigate mitochondrial damage. -
表 1 引物序列
Table 1. Primer sequences
基因 序列(5’→ 3’) XBP1s 正义链 AAGAACACGCTTG GGAATGG 反义链 CTGCACCTGCTGCGGAC ITPR1 正义链 CTCTGTATGCGGAGGGATCTAC 反义链 CAGGATACTTAGCTATGAGGCG ITPR2 正义链 CTTCCTCTACATTGGGGACATC 反义链 GGGATACTTAGCTATGAGACGG ITPR3 正义链 CGACCTCATCAAGGCTCTCC 反义链 TCCCCATCTCGTTGTTCTGC β-actin 正义链 AGGCCAACCGTGAAAGATG 反义链 TGGCGTGAGGGAGAGCATAG -
[1] THUILLIER R. Molecular frontiers in transplantation-induced ischemia-reperfusion injury[J]. Int J Mol Sci, 2023, 24(4): 3450. DOI: 10.3390/ijms24043450. [2] KELLUM JA, ROMAGNANI P, ASHUNTANTANG G, et al. Acute kidney injury[J]. Nat Rev Dis Primers, 2021, 7(1): 52. DOI: 10.1038/s41572-021-00284-z. [3] ZHANG J, WEI X, TANG Z, et al. Elucidating the molecular pathways and immune system transcriptome during ischemia-reperfusion injury in renal transplantation[J]. Int Immunopharmacol, 2020, 81: 106246. DOI: 10.1016/j.intimp.2020.106246. [4] DERY KJ, KUPIEC-WEGLINSKI JW. New insights into ischemia-reperfusion injury signaling pathways in organ transplantation[J]. Curr Opin Organ Transplant, 2022, 27(5): 424-433. DOI: 10.1097/MOT.00000000000 01005. [5] NIEUWENHUIJS-MOEKE GJ, PISCHKE SE, BERGER SP, et al. Ischemia and reperfusion injury in kidney transplantation: relevant mechanisms in injury and repair[J]. J Clin Med, 2020, 9(1): 253. DOI: 10.3390/jcm9010253. [6] 叶芃, 程帆. 线粒体在肾脏缺血再灌注损伤中的作用与机制[J]. 医学综述, 2022, 28(4): 643-648. DOI: 10.3969/j.issn.1006-2084.2022.04.004.YE P, CHENG F. Role and mechanism of mitochondria in renal ischemia-reperfusion injury[J]. Med Recap, 2022, 28(4): 643-648. DOI: 10.3969/j.issn.1006-2084.2022.04.004. [7] CHEN Q, KOVILAKATH A, ALLEGOOD J, et al. Endoplasmic reticulum stress and mitochondrial dysfunction during aging: role of sphingolipids[J]. Biochim Biophys Acta Mol Cell Biol Lipids, 2023, 1868(10): 159366. DOI: 10.1016/j.bbalip.2023.159366. [8] 于露, 李永华. 线粒体相关内质网膜的生物学功能及其在相关疾病中作用的研究进展[J/CD]. 中华诊断学电子杂志, 2022, 10(4): 284-288. DOI: 10.3877/cma.j.issn.2095-655X.2022.04.014.YU L, LI YH. Research progress on biological function of mitochondria-associated endoplasmic reticulum membranes and its role in related diseases[J/CD]. Chin J Diagn (Electr Edit), 2022, 10(4): 284-288. DOI: 10.3877/cma.j.issn.2095-655X.2022.04.014. [9] GAO P, YANG W, SUN L. Mitochondria-associated endoplasmic reticulum membranes (MAMs) and their prospective roles in kidney disease[J]. Oxid Med Cell Longev, 2020: 3120539. DOI: 10.1155/2020/3120539. [10] WOLL KA, VAN PETEGEM F. Calcium-release channels: structure and function of IP3 receptors and ryanodine receptors[J]. Physiol Rev, 2022, 102(1): 209-268. DOI: 10.1152/physrev.00033.2020. [11] YUAN L, LIU Q, WANG Z, et al. EI24 tethers endoplasmic reticulum and mitochondria to regulate autophagy flux[J]. Cell Mol Life Sci, 2020, 77(8): 1591-1606. DOI: 10.1007/s00018-019-03236-9. [12] YUAN M, GONG M, HE J, et al. IP3R1/GRP75/VDAC1 complex mediates endoplasmic reticulum stress-mitochondrial oxidative stress in diabetic atrial remodeling[J]. Redox Biol, 2022, 52: 102289. DOI: 10.1016/j.redox.2022.102289. [13] ZIEGLER DV, VINDRIEUX D, GOEHRIG D, et al. Calcium channel ITPR2 and mitochondria-ER contacts promote cellular senescence and aging[J]. Nat Commun, 2021, 12(1): 720. DOI: 10.1038/s41467-021-20993-z. [14] MORCIANO G, GIORGI C, BONORA M, et al. Molecular identity of the mitochondrial permeability transition pore and its role in ischemia-reperfusion injury[J]. J Mol Cell Cardiol, 2015, 78: 142-153. DOI: 10.1016/j.yjmcc.2014.08.015. [15] NI H, OU Z, WANG Y, et al. XBP1 modulates endoplasmic reticulum and mitochondria crosstalk via regulating NLRP3 in renal ischemia/reperfusion injury[J]. Cell Death Discov, 2023, 9(1): 69. DOI: 10.1038/s41420-023-01360-x. [16] ZHANG J, ZHANG J, NI H, et al. Downregulation of XBP1 protects kidney against ischemia-reperfusion injury via suppressing HRD1-mediated NRF2 ubiquitylation[J]. Cell Death Discov, 2021, 7(1): 44. DOI: 10.1038/s41420-021-00425-z. [17] LIU H, WANG L, WENG X, et al. Inhibition of Brd4 alleviates renal ischemia/reperfusion injury-induced apoptosis and endoplasmic reticulum stress by blocking FoxO4-mediated oxidative stress[J]. Redox Biol, 2019, 24: 101195. DOI: 10.1016/j.redox.2019.101195. [18] SMITH SF, HOSGOOD SA, NICHOLSON ML. Ischemia-reperfusion injury in renal transplantation: 3 key signaling pathways in tubular epithelial cells[J]. Kidney Int, 2019, 95(1): 50-56. DOI: 10.1016/j.kint.2018.10.009. [19] KUMAR V, MAITY S. ER Stress-sensor proteins and ER-mitochondrial crosstalk-signaling beyond (ER) stress response[J]. Biomolecules, 2021, 11(2): 173. DOI: 10.3390/biom11020173. [20] HUANG R, HUI Z, WEI S, et al. IRE1 signaling regulates chondrocyte apoptosis and death fate in the osteoarthritis[J]. J Cell Physiol, 2022, 237(1): 118-127. DOI: 10.1002/jcp.30537. [21] DE RIDDER I, KERKHOFS M, LEMOS FO, et al. The ER-mitochondria interface, where Ca2+ and cell death meet[J]. Cell Calcium, 2023, 112: 102743. DOI: 10.1016/j.ceca.2023.102743. [22] HULSURKAR MM, LAHIRI SK, KARCH J, et al. Targeting calcium-mediated inter-organellar crosstalk in cardiac diseases[J]. Expert Opin Ther Targets, 2022, 26(4): 303-317. DOI: 10.1080/14728222.2022.2067479. [23] BASSO V, MARCHESAN E, ZIVIANI E. A trio has turned into a quartet: DJ-1 interacts with the IP3R-GRP75-VDAC complex to control ER-mitochondria interaction[J]. Cell Calcium, 2020, 87: 102186. DOI: 10.1016/j.ceca.2020.102186. [24] 徐王婷, 宋育林. 抑制IP3R-Ca2+途径对对乙酰氨基酚所致肝损伤及其线粒体内质网结构偶联的影响[J]. 安徽医科大学学报, 2023, 58(7): 1077-1081. DOI: 10.19405/j.cnki.issn1000-1492.2023.07.003.XU WT, SONG YL. The effects of inhibition of the IP3R-Ca2+pathway in APAP-induced liver injury and mitochondrial-associated endoplasmic reticulum membranes[J]. Acta Univ Med Anhui, 2023, 58(7): 1077-1081. DOI: 10.19405/j.cnki.issn1000-1492.2023.07.003. [25] PENNA E, ESPINO J, DE STEFANI D, et al. The MCU complex in cell death[J]. Cell Calcium, 2018, 69: 73-80. DOI: 10.1016/j.ceca.2017.08.008. [26] 史喜德, 郭珊珊. MICU1及其与相关疾病的作用机制[J]. 临床与病理杂志, 2022, 42(7): 1737-1744. DOI: 10.3978/j.issn.2095-6959.2022.07.033.SHI XD, GUO SS. Mechanism of MICU1 and its related diseases[J]. J Clin Pathol Res, 2022, 42(7): 1737-1744. DOI: 10.3978/j.issn.2095-6959.2022.07.033. [27] ARRUDA AP, PERS BM, PARLAKGÜL G, et al. Chronic enrichment of hepatic endoplasmic reticulum-mitochondria contact leads to mitochondrial dysfunction in obesity[J]. Nat Med, 2014, 20(12): 1427-1435. DOI: 10.1038/nm.3735. [28] CHU B, LI M, CAO X, et al. IRE1α-XBP1 affects the mitochondrial function of Aβ25-35-treated SH-SY5Y cells by regulating mitochondria-associated endoplasmic reticulum membranes[J]. Front Cell Neurosci, 2021, 15: 614556. DOI: 10.3389/fncel.2021.614556. [29] HAJHEIDARI M, HUANG SC. Elucidating the biology of transcription factor-DNA interaction for accurate identification of cis-regulatory elements[J]. Curr Opin Plant Biol, 2022, 68: 102232. DOI: 10.1016/j.pbi.2022.102232. [30] INUKAI S, KOCK KH, BULYK ML. Transcription factor-DNA binding: beyond binding site motifs[J]. Curr Opin Genet Dev, 2017, 43: 110-119. DOI: 10.1016/j.gde.2017.02.007. [31] WANG P, SU J, WANG J, et al. NRF1 promotes primordial germ cell development, proliferation and survival[J]. Cell Prolif, 2024, 57(1): e13533. DOI: 10.1111/cpr.13533. [32] WANG Z, COBAN B, WU H, et al. GRHL2-controlled gene expression networks in luminal breast cancer[J]. Cell Commun Signal, 2023, 21(1): 15. DOI: 10.1186/s12964-022-01029-5. [33] ZHANG X, ZHANG C, QIAO M, et al. Depletion of BATF in CAR-T cells enhances antitumor activity by inducing resistance against exhaustion and formation of central memory cells[J]. Cancer Cell, 2022, 40(11): 1407-1422. DOI: 10.1016/j.ccell.2022.09.013. [34] 颜凤, 邱佳韵, 庄丽丽, 等. 转录因子STAT1对癌症相关基因Gas6的调控研究[J]. 南京医科大学学报(自然科学版), 2023, 43(11): 1487-1493. DOI: 10.7655/NYDXBNS20231102.YAN F, QIU JY, ZHUANG LL, et al. The regulation of the transcription factor STAT1 to cancer-related gene Gas6[J]. J Nanjing Med Univ(Nat Sci), 2023, 43(11): 1487-1493. DOI: 10.7655/NYDXBNS20231102. [35] 许晓亮, 刘辉, 武燃, 等. 缺氧诱导因子诱导转录因子COUP-TFⅡ转录激活[J]. 中国老年学杂志, 2022, 42(21): 5298-5303. DOI: 10.3969/j.issn.1005-9202.2022.21.043.XU XL, LIU H, WU R, et al. Hypoxia inducible factor induces transcriptional activation of transcription factor COUP-TF II[J]. Chin J Gerontol, 2022, 42(21): 5298-5303. DOI: 10.3969/j.issn.1005-9202.2022.21.043. [36] HUA C, HUANG J, WANG T, et al. Bacterial transcription factors bind to coding regions and regulate internal cryptic promoters[J]. mBio, 2022, 13(5): e0164322. DOI: 10.1128/mbio.01643-22. [37] CHEN H, PUGH BF. What do transcription factors interact with?[J]. J Mol Biol, 2021, 433(14): 166883. DOI: 10.1016/j.jmb.2021.166883. [38] ANDREWS G, FAN K, PRATT HE, et al. Mammalian evolution of human cis-regulatory elements and transcription factor binding sites[J]. Science, 2023, 380(6643): eabn7930. DOI: 10.1126/science.abn7930. [39] KRIEGER G, LUPO O, WITTKOPP P, et al. Evolution of transcription factor binding through sequence variations and turnover of binding sites[J]. Genome Res, 2022, 32(6): 1099-1111. DOI: 10.1101/gr.276715.122. [40] GEORGAKOPOULOS-SOARES I, DENG C, AGARWAL V, et al. Transcription factor binding site orientation and order are major drivers of gene regulatory activity[J]. Nat Commun, 2023, 14(1): 2333. DOI: 10.1038/s41467-023-37960-5. [41] WEI S, LI X, LU Z, et al. A transcriptional regulator that boosts grain yields and shortens the growth duration of rice[J]. Science, 2022, 377(6604): eabi8455. DOI: 10.1126/science.abi8455. [42] DU Y, LIU L, PENG Y, et al. UNBRANCHED3 expression and inflorescence development is mediated by UNBRANCHED2 and the distal enhancer, KRN4, in maize[J]. PLoS Genet, 2020, 16(4): e1008764. DOI: 10.1371/journal.pgen.1008764. -




 下载:
下载: